Pattern matching starts by defining a template, in our case, a
tumour-like template. The definition of the template is based on
the approach of Lai et al. [98], who defined the tumour by
three characteristics: brightness contrast, uniform density and
circular shape. In our implementation, the template can vary
between ![]() and
and ![]() pixels in diameter.
Figure
pixels in diameter.
Figure ![]() shows a
shows a ![]() -pixel radius
template. The circular patch of ones in the centre represents a
tumour area having uniform density. The ring of zeros represents
the ``don't care" area to account for some of the shape
variability. Finally, the outer edge of the template is filled
with minus ones to represent the dark background. One of the
drawbacks of this algorithm is its poor performance in detecting
spiculated masses [98].
-pixel radius
template. The circular patch of ones in the centre represents a
tumour area having uniform density. The ring of zeros represents
the ``don't care" area to account for some of the shape
variability. Finally, the outer edge of the template is filled
with minus ones to represent the dark background. One of the
drawbacks of this algorithm is its poor performance in detecting
spiculated masses [98].
In contrast with the original work, where the authors used a cross-correlation metric to measure the similarity among the image patches and the template, in this work we used a mutual information based metric. This similarity measure was inspired on the work of Tourassi et al. [179], where they used it to retrieve similar RoIs in a CBIR system. As shown in [129], the results obtained using this probabilistic metric outperforms the ones obtained using the cross-correlation metric.
Given two images A and B, the mutual information is expressed as:
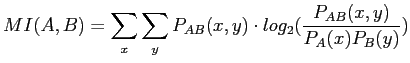 |
(2.12) |
where
![]() is the joint probability of the two
images based on their corresponding pixels values and
is the joint probability of the two
images based on their corresponding pixels values and ![]() and
and
![]() are the marginal probabilities of the variables
are the marginal probabilities of the variables ![]() and
and
![]() which are the image pixel values, and are obtained from the
corresponding normalized histograms. To obtain a compatible
template we calculated the mean of all pixels in the breast.
Subsequently, in the template,
which are the image pixel values, and are obtained from the
corresponding normalized histograms. To obtain a compatible
template we calculated the mean of all pixels in the breast.
Subsequently, in the template, ![]() 's are replaced with pixels
values inferiors to the mean, 0
's with the value of the mean,
and
's are replaced with pixels
values inferiors to the mean, 0
's with the value of the mean,
and ![]() with values superiors to it.
with values superiors to it.